The goal was to provide a software beeing able to compute hepatic tumoral areas in a semi automatic way. The software would compute the area on input images, and allow the detected areas to be manually modified to correct potential misdetections.
The input data are sets of X-rays pictures, exported from DICOM to jpeg. A set of input data is usually between 15 and 30 pictures, but this is not an important factor, as full analysis is performed on each image, without any use of data from other images.
This section will explain the algorithm used to perform the hepatic tumoral area segmentation.
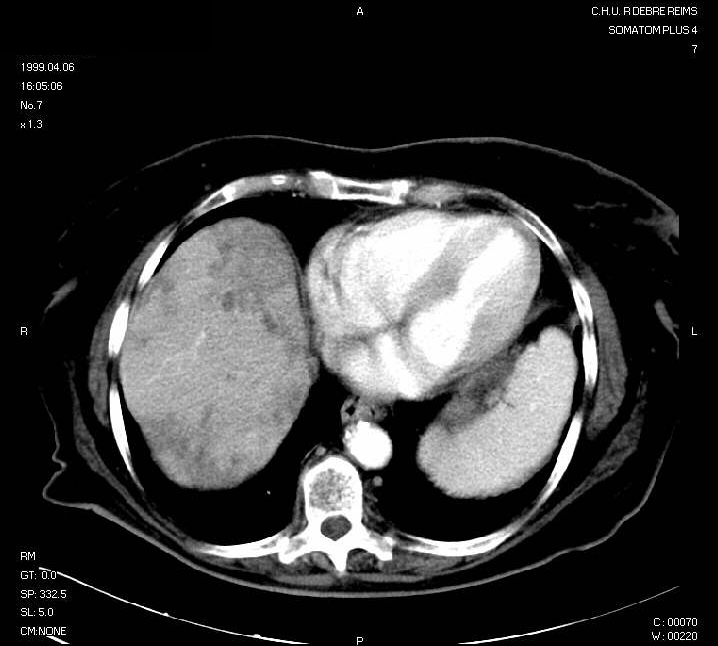
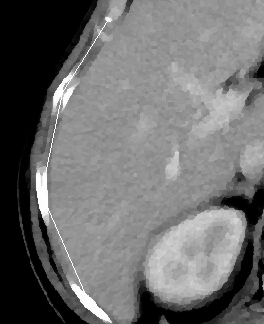
Here is an example of (reduced) input picture:

This example picture will be used as an example. The hepatic area is on the left side. Darker areas are tumoral areas.
Because of the noise in the input image, some noise removal will be used. A Kuwahara filter has been used.
Before Kuwahara filtering (enlarged):

After Kuwahara filtering (enlarged):

After Kuwahara filtering:
An interesting point about this filtering is that it also enhance edges.
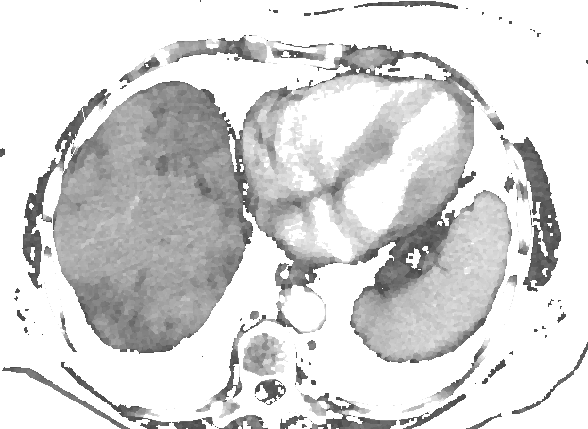
Bones that are near the area of interest are then detected.
Bones:
Bones after detection:
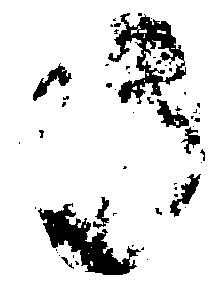
A thresholding allows to keep only the parts of the input picture that are of a specific grey level range.
Thresholded image:
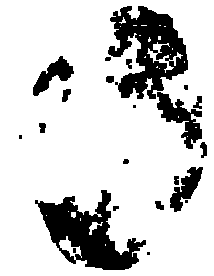
A cross-shaped neighbourhood is used on the picture. If there is at least one white pixel inside the neighbourhood of a pixel, this one is removed.
Small areas are then removed:

The biggest area is kept. This one is the hepatic area.
Segmented hepatic area:

Borders are removed:

An adaptative thresholding is performed in order to identify tumoral areas.
Result:
At this step, we have a lot of small tumoral areas detected. A dilatation is performed in order to join them.
Dilatation:
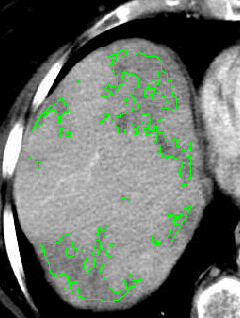
We now have successfully detected the hepatic tumoral area on this X-Ray picture. Here is the result: